Co-dominant SCAR Markers for Detection of the Ty-3 and Ty-3a Loci from Solanum chilense
at 25 cM of Chromosome 6 of Tomato
Yuanfu Ji1, Melinda S. Salus2, Bram van Betteray3, Josie Smeets3, Katie S. Jensen2, Christopher T. Martin2, Luis Mejía4, Jay W. Scott1, Michael J. Havey5, and Douglas P. Maxwell2
1University of Florida, IFAS, Gulf Coast Research & Education Center, 14625 CR 672, Wimauma, FL 33598
2University of Wisconsin-Madison, Dept. of Plant Pathology, 1630 Linden Dr., Madison, WI 53706
3Nunhems BV, PO Box 4005, 6080 AC Haelen, The Netherlands
4Universidad de San Carlos, Ciudad de Guatemala Zona 12, Guatemala
5University of Wisconsin-Madison, Dept. of Horticulture, Madison, WI and U.S. Dept. Agr.
Email: dpmax@plantpath.wisc.edu
Breeding for resistance to begomoviruses in tomato can be greatly aided by the availability of PCR-based markers for the various resistance loci. Four begomovirus-resistance loci or regions have been mapped to chromosome 6 (Agrama and Scott, 2006; Chagué et al., 1997; Ji and Scott, 2006b; Ji et al., 2007; Zamir et al., 1994). The Ty-1 locus, which is part of the introgression derived from Solanum chilense LA1969, is located between markers TG297 (4 cM) and TG97 (8.6 cM) (Zamir et al., 1994). Agrama and Scott (2006) reported three regions that contributed to resistance in breeding lines with introgressions from S. chilense LA2779 or LA1932. One region corresponded to the region having the Ty-1 locus. Another region was the Ty-3 locus, which was mapped to a region between cLEG-31-P16 (20 cM) and T1079 (27 cM) (Ji and Scott, 2006b; Ji et al., 2007). The third region was near the self-pruning (sp) and potato leaf (c) loci. Another begomovirus-resistance QTL, derived from an introgression from S. pimpinellifolium, was mapped near the marker TG153 (33 cM; Chagué et al., 1997).
Previously, Ji and Scott (2006a, 2006b; Ji et al., 2007) reported the development of SCAR and CAPS markers linked to begomovirus resistance genes derived from S. chilense on chromosome 6, and they determined that the Ty-3 locus mapped to a region that included the FER locus (25 cM, BAC clone 56B23, AY678298). Jensen and Maxwell (Maxwell et al., 2007) found that the sequences for the G8 gene of the BAC clone 56B23 are different for lines derived from LA2779 and LA1932. To differentiate the two introgressions, the one from LA2779 is designated Ty-3 and the one from LA1932, Ty-3a. This report describes two sets of PCR primers that provide co-dominant SCAR markers for detection of the Ty-3 and Ty-3a introgressions.
Primer Design:
Initially, PCR primers were designed to amplify sequences near the 5’ end of the BAC clone 56B23. These primers were used to amplify PCR fragments from the begomovirus-susceptible heritage tomato, S. lycopersicum ‘Purple Russian’, and a begomovirus-resistant breeding line, Gc43-3, from the tomato breeding program at San Carlos University, Guatemala with an introgression in this region from S. chilense LA2779 (Mejía et al., 2005). These sequences were aligned, and forward and reverse primers designed from conserved regions: forward primer FLUW-25F (5’ CAAGTGTGCATATACTTCATA(T/G)TCACC) and reverse primer, FLUW-25R (5’ CCATATATAACCTCTGTTTCTATTTCGAC). As expected, these primers gave PCR fragments for S. lycopersicum and the S. chilense introgression of 475 and 641 bp, respectively. A third primer pair was designed to give smaller PCR fragments from the aligned sequences of the FLUW25 fragment for the forward primer and sequences 3’ of the FLUW-25R primer for the reverse primer: forward primer, P6-25-F2, 5’ GGT AGT GGA AAT GAT GCT GCT C, and reverse primer, P6-25-R5, 5’ GCT CTG CCT ATT GTC CCA TAT ATA ACC. The P6-25-F2/P6-25-R5 primers were expected to give fragment sizes for S. lycopersicum and the S. chilense LA2779 introgression of 320 bp and 453 bp, respectively.
DNA Extraction and PCR Methods: DNA was extracted from fresh leaves of plants with PUREGENE® DNA Purification Kit (Gentra Systems, Inc., Minneapolis, MN) and DNA adjusted to approximately 10 ng/µl. PCR parameters were for 25-µl reactions containing 2.5 µl 2.5 mM dNTPs, 5 µl 5x buffer, 2.5 µl 2.5 mM MgCl2, 0.1 µl (0.5 units) GoTaq DNA polymerase (Promega Corp., Madison, WI), 2.5 µl each forward and reverse primer at 10 μM, 2-5 µl of DNA extract, and water. PCR cycles were 94 C for 4 min, 35 cycles of 94 C for 30 sec, 53 C for 1 min, and 72 C for 1 min. These cycles were followed by 72 C for 10 min, and then the reaction was held at 4 C. PCR reactions were performed in the MJ DNA Engine PT200 Thermocycler™ (MJ Research Inc., Waltham, MA). PCR-amplified fragments were separated by gel electrophoresis with 1.5% agarose in 0.5 X TBE buffer, stained with ethidium bromide, and visualized with UV light. ssDNA was digested in PCR reactions with shrimp alkaline phosphatase (Promega Corp.) and exonuclease I (Epicentre, Madison, WI) and the PCR-fragments were directly sequenced with Big Dye Sequencing Kit™ and analyzed by the Biotechnology Center, University of Wisconsin-Madison.
Results and Discussion:
The FLUW-25 primer pair amplified fragments of 480 bp and 640 bp from the susceptible line S. lycopersicum Heinz 1706 and from the begomovirus-resistant breeding line, Gc43-3, with an introgression from S. chilense LA2779, respectively. Surprisingly, the Gh25-3 breeding line, which was derived from the begomovirus-resistant line Ih902 (see line 902 in Vidavsky and Czosnek, 1998) with resistance reported to be from an introgression from S. habrochaites, also gave a 640-bp fragment. The heterozygous plant, Gh228-1, with resistance from Ih902, gave the two sizes, 480 and 640 bp, for the Ty-3/ty-3 genotype (Fig. 1).
1 2 3 4 5 6
![]()
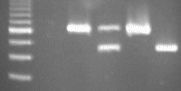
Fig. 1. PCR fragments with primers FLUW25F/FLUW25R; Lane 1, 100-bp marker, Invitrogen, bright band, 600 bp; Lane 2, water control; Lane 3, Gc43-3 (resistant); Lane 4, Gh228-1 (resistant, heterozygous); Lane 5, Gh25-3 (resistant); Lane 6, Heinz 1706 (susceptible).
The sequences for the FLUW25-PCR fragments from Heinz (475 bp) and Gc43-3 (641 bp) were aligned; and there were 18 SNPs and 5 indel differences (Maxwell et al., 2007). Single indels of 3, 6, 42, and 143 nt and two indels of 5 nt were present. The sequence for the fragment from Heinz had 100% nt identity with the sequence of the comparable region of the BAC clone 56B23. In another susceptible line, M82-1-8, the sequence for the 475-bp fragment was identical to that of Heinz. Surprisingly, the sequence for the 640-bp fragment from Gh25-3 with resistance from Ih902 had 100% nt identity with the fragment from Gc43-3. The two fragments from Gh228-1 had 100% nt identity with the 475- and 641-bp fragments from Heinz and Gc43-3, respectively. Additionally, the sequence of the 640-bp fragments from 11 other begomovirus-resistant breeding lines with resistance from either S. chilense LA2779 or Ih902 had 99-100% nt identity with Gc43-3. These FLUW25 primers were used to evaluate the presence of the Ty-3 locus in 102 breeding lines from the tomato breeding program at San Carlos University, Guatemala (Mejía et al., 2005), and the results were as expected except for the Gc171 line. This line was known to have an introgression, Ty-3a, in this region derived from S. chilense LA1932, but no PCR fragment was amplified by these primers. van Betteray and Smeets (unpublished) determined that the FLUW25R primer did not anneal to S. chilense LA1932 sequences. Thus, several more reverse primers were designed from the same area of the BAC clone 56B23 sequence. These were evaluated with Gc171 or lines selected from Gc171 crosses. One primer, P6-25-R5, gave fragments with FLUW25F for S. chilense LA1932 derived lines.
An additional primer set, P6-25-F2 and P6-25-R5, was designed to include the 143-nt ty-3/Ty-3 indel and to give smaller fragments than the FLUW25 primer set (Fig. 2). With begomovirus-resistant breeding lines derived from either the S. chilense LA2779 source, Gc9, or the Ih902 line (Vidavsky and Czosnek, 1998), the expected 450-bp Ty-3 fragment was obtained. A 320-bp ty-3 fragment was amplified from breeding lines lacking the introgression from either of these two begomovirus-resistance sources. A 630-bp Ty-3a fragment was obtained from lines derived from S. chilense LA1932, such as Gc171. Heterozygous hybrids were easily detected with these primers which amplified two fragments corresponding to the S. lycopersicum ty-3 fragment (320 bp) and either the Ty-3 (450 bp) or the Ty-3a (630 bp) fragment (Fig. 2). No F1 hybrids were available to test for fragments with the Ty-3/Ty-3a genotype, but it is expected that this primer pair would also amplify two fragments (450 and 630 bp) with this genotype.
1 2 3 4 5 6 7
![]()
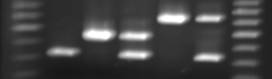
Fig. 2. PCR fragments with primers P6-25-F2/P6-25-R5. Lane 1, 100-bp Brenchtop DNA ladder, Promega; Lane 2, M82-1-8 (ty-3/ty-3); Lane 3, Gc9 (Ty-3/Ty-3); Lane 4, Romelia, F1 hybrid, (Ty-3/ty-3); Lane 5, Gc171 (Ty-3a/Ty-3a); Lane 6, GTc191-3, F1 hybrid, (Ty-3a/ty-3); Lane 7, 100-bp marker.
The three sizes of the P6-25-F2/P6-25-R5 fragments were sequenced (Maxwell et al., 2007). The 320-bp and the 450-bp fragments corresponded to the sequences of S. lycopersicum and of the Ty-3 locus associated with lines derived from S. chilense LA2779, respectively. The 650-bp fragment from Gc171 had two large inserts, when compared with the S. lycopersicum sequence.
Conclusions: These two sets of primers detect co-dominant SCAR markers, FLUW25 and P6-25, for the ty-3, Ty-3 and Ty-3a loci. It is not known how closely these markers are to the functional Ty-3 gene (Ji et al., 2007), so it is possible that some breeding lines would give false negative or false positive results. It is expected that these markers will be evaluated in various tomato breeding programs.
Acknowledgements: This project was funded in part by USAID-CDR (TA-MOU-05-C25-037) and USAID-MERC (GEG—G-00-02-00003-00) grants to D. P. Maxwell, by the College of Agricultural and Life Sciences at University of Wisconsin-Madison, and by grants from Unilever Bestfoods Ltd. and the Florida Tomato Committee to J. W. Scott.
Literature Cited:
Agrama, H.A., and Scott, J.W. 2006. Quantitative trait loci for tomato yellow leaf curl virus and tomato mottle virus resistance in tomato. J. Am. Hortic. Sci. 131:267-272.
Chagué, V., Mercier, J.C., Guenard, M., de Courcel, A., and Vedel, F. 1997. Identification of RAPD markers linked to a locus involved in quantitative resistance to TYLCV in tomato by bulked segregant analysis. Theor. Appl. Genet. 95:671-677.
Ji, Y., and Scott, J.W. 2006a. Development of breeder friendly markers for begomovirus resistance genes derived from L. chilense. Proc Tomato Breeders Table, Tampa, FL, USA. roundtable06.ifas.ufl.edu/Schedule.htm
Ji, Y. and Scott, J.W. 2006b. Ty-3, a begomovirus resistance locus linked to Ty-1 on chromosome 6. Rept. Tomato Genetics Coop. 56:22-25.
Ji, Y., Schuster, D.J., and Scott, J.W. 2007. Ty-3, a begomovirus resistance locus near the Tomato yellow leaf curl virus resistance locus Ty-1 on chromosome 6 of tomato. Mol. Breeding 20:271-284
Maxwell, D.P., Martin, C.T., Garcia, B.E., Salus, M.S., Jensen, K.S, Havey, M.J. and Mejia, L. 2007. Markers for tomato chromosomes. www.plantpath.wisc.edu/GeminivirusResistantTomatoes
Mejía, L., Teni, R.E., Vidavski, F., Czosnek, H., Lapidot, M., Nakhla, M.K., and Maxwell, D.P. 2005. Evaluation of tomato germplasm and selection of breeding lines for resistance to begomoviruses in Guatemala. Acta Hort. 695:251-255.
Vidavsky, F., and Czosnek, H. 1998. Tomato breeding lines immune and tolerant to tomato yellow leaf curl virus (TYLCV) issued from Lycopersicon hirsutum. Phytopathology 88:910-914.
Zamir, D., Michelson, I., Zakay, Y., Navot, N., Zeidan, N., Sarfatti, M., Eshed, Y., Harel, E., Pleban, T., van-Oss, H., Kedar, N., Rabinowitch, H.D., and Czosnek, H. 1994. Mapping and introgression of a tomato yellow leaf curl virus tolerance gene, Ty-1. Theor. Appl. Genet. 88:141-146.