Ty-3, a begomovirus resistance locus linked to Ty-1 on chromosome 6 of tomato
Yuanfu Ji and J. W. Scott
University of Florida, IFAS, Gulf Coast Research & Education Center, 14625 CR 672, Wimauma, FL 33598, USA email: yji@ufl.edu, jwsc@ufl.edu
Whitefly-transmitted begomoviruses, such as tomato yellow leaf curl virus (TYLCV) and tomato mottle virus (ToMoV), cause great losses to tomato production in the United States and other tropical and subtropical regions in the world (Polston and Anderson, 1997). To date, no resistance has been found in the Solanum lycopersicum germplasm although some tomato varieties have been reported to be less susceptible than others during severe epidemics (Hassan et al., 1991, Laterrot, 1993). However, resistance to TYLCV has been found in numerous tomato wild species, including S. pimpinellifolium, S. peruvianum, S. chilense, S. habrochaites, and S. cheesmaniae (Pico et al., 1996). S. chilense accession LA1969 showed the highest level of resistance among the 23 accessions representing 5 tomato species based on symptom expression and virus detection criteria (Zakay et al., 1991). A partially dominant major gene, Ty-1, which contributed most of the resistance to TYLCV in LA1969, was mapped closely to the RFLP marker TG97 on chromosome 6 (Zamir et al., 1994). Another gene locus responsible for resistance to TYLCV in the tomato line ‘H24” derived from S. habrochaites was localized to the long arm of chromosome 11 delimited by RFLP markers TG393 and TG36 (Hanson et al., 2000). This locus was further delimited to a smaller interval (P. Hanson, personal communication), and formally named as Ty-2 (Hanson et al., 2006).
S. chilense accessions LA1932, LA2779 and LA1938 also showed high levels of resistance to begomoviruses (Scott and Schuster, 1991, Scott et al., 1996) and have been useful sources of resistance in the tomato breeding program in Florida and elsewhere in the world (Mejía et al., 2005, Scott, 2001). Inheritance studies and QTL mapping analysis using RAPD markers revealed three regions on chromosome 6 contributing to resistance to both TYLCV and ToMoV in these accessions (Agrama and Scott, 2006, Griffiths, 1998). The first region encompass the Ty-1 region, while the other two regions flank either side of the self-pruning (sp) and potato leaf (c) loci. The RAPD markers mapped to these resistance regions were used to screen more advanced resistant breeding lines in search for tightly linked markers, which were then converted to sequence characterized amplified region (SCAR) markers (Ji and Scott, 2005a, b).
These SCAR markers, as well as other PCR-based markers on chromosome 6 obtained from public domains or designed from public sequences, were used in the present study to localize the introgression in the advanced breeding lines derived from LA2779, which were resistant to both TYLCV and ToMoV. A large introgressed segment was found in these lines, which spans markers from C2_At2g39690 at 5.3 cM in Tomato-EXPEN 2000 map (Fulton et al., 2002; http://sgn.cornell.com) to T0834 (32 cM) (Figure 1). Using an F2 population of susceptible S. lycopersicum ´ a resistant advanced breeding line having this introgression, we mapped a partially dominant gene, that we hereby designate Ty-3, to the marker interval between cLEG-31-P16 (20 cM) and T1079 (27 cM) on the long arm of chromosome 6. This gene has a dominance-to-additive effect ratio of 0.47 for resistance to TYLCV, suggesting a nearly equal contribution to the variance in TYLCV resistance from additive and dominance effects. Additionally, ~65% of the variance in TYLCV resistance in the F2 progeny can be explained from this gene locus, suggesting this gene had a major effect on the resistance. Besides TYLCV resistance, Ty-3 might also contribute resistance to ToMoV, but to a lesser degree, although the possibility of a different gene locus in the same region accounting for ToMoV resistance cannot be ruled out. QTL analysis of F2 progeny from the same cross, but inoculated with ToMoV, revealed that ~41% of the variance in ToMoV resistance in the progeny could be explained by this gene locus, which had a dominance-to-additive ratio of 0.35. No recombinants were found between TG590 (22 cM) and T1079, which prevented a fine-scale mapping of the Ty-3 gene using this LA2779-derived F2 population.
Besides the Ty-3 gene, introgression in the advanced breeding lines derived from LA2779 also span the Ty-1 region near the Mi gene, suggesting possible coexistence and linkage of resistance alleles at both Ty-1 and Ty-3 loci in these lines. The large introgression in these breeding lines was further confirmed from sequence alignments at numerous marker loci, including markers located in both Ty-1 and Ty-3 regions (Maxwell et al., 2006). At the REX-1 locus, which is tightly linked to the Ty-1 gene (Milo, 2001), the sequence of a LA2779-derived advanced breeding line (Gc9) was identical to that of TY52, a TYLCV-resistance line that is homozygous for Ty-1 gene (Maxwell et al., 2006, Zamir et al., 1994). The presence of both Ty-1 and Ty-3 genes in a single genotype most likely offers the highest resistance to TYLCV, which might be the case for some of the present commercial hybrids in the market (Ji and Scott, unpublished data).
Literature Cited:
Agrama, H. A. and J. W. Scott. 2006. Quantitative trait loci for tomato yellow leaf curl virus and tomato mottle virus resistance in tomato. J. Am. Soc. Hort. Sci. 131:267-272.
Fulton, T. M., R. Van der Hoeven, N. T. Eannetta, and S. D. Tanksley. 2002. Identification, analysis, and utilization of conserved ortholog set markers for comparative genomics in higher plants. Plant Cell 14:1457-1467.
Griffiths, P. D. 1998. Inheritance and linkage of geminivirus resistance genes derived from Lycopersicon chilense (Dunal) in tomato (Lycopersicon esculentum Mill). Ph.D Dissertation, University of Florida, Gainesville.
Hanson, P., S. K. Green, and G. Kuo. 2006. Ty-2, a gene on chromosome 11 conditioning geminivirus resistance in tomato. Tomato Genet. Coop. Rep. 56:17-18.
Hanson, P. M., D. Bernacchi, S. Green, S. D. Tanksley, Muniyappa V, P. AS, and e. al. 2000. Mapping a wild tomato introgression associated with tomato yellow leaf curl virus resistance in a cultivated tomato line. J. Am. Soc. Hort. Sci. 15:15–20.
Hassan, A. A., M. S. Wafi, N. E. Quronfilah, U. A. Obaji, M. A. Al-Rayis, and F. Al-Izabi. 1991. Screening for tomato yellow leaf curl virus resistance in wild and domestic Lycopersicon accessions. Tomato Genet. Coop. Rep. 41:19-21.
Ji, Y. and J. W. Scott. 2005a. Identification of RAPD markers linked to Lycopersicon chilense derived geminivirus resistance genes on chromosome 6 of tomato. Acta Hort. 695:407-416.
Ji, Y. and J. W. Scott. 2005b. Development of SCAR and CAPS markers linked to tomato begomovirus resistance genes introgressed from Lycopersicon chilense. HortSci. 40:1090.
Stamova, L. (ed.) 1993. Proceedings of the Xllth Eucarpia Meeting on Tomato Genetics and Breeding, Plovdiv, Bulgaria. 27-31 July. Maritsa Vegetable Crops Research Institute, Plovdiv, Bulgaria.
Maxwell, D. P., C. Martin, M. S. Salus, L. Montes, and L. Mejía. 2006. Tagging begomovirus resistance genes. www.plantpath.wisc.edu/GeminivirusResistantTomatoes/Markers.
Mejía, L., R. E. Teni, F. Vidavski, H. Czosnek, M. Lapidot, M. K. Nakhla, and D. P. Maxwell. 2005. Evaluation of tomato germplasm and selection of breeding lines for resistance to begomoviruses in Guatemala. Acta Hort. 695:251-256.
Milo, J. 2001. The PCR-based marker REX-1, linked to the gene Mi, can be used as a marker to TYLCV tolerance, in Proceedings of Tomato Breeders Roundtable, Antigua, Guatemala, www.oardc.ohio-state.edu/tomato/TBRT%202001%20Abstracts.pdf.
Pico, B., M. J. Diez, and F. Nuez. 1996. Viral diseases causing the greatest economic losses to the tomato crop .2. The tomato yellow leaf curl virus - A review. Sci. Hort.-Amsterdam 67:151-196.
Polston, J. E. and P. K. Anderson. 1997. The emergence of whitefly-transmitted geminiviruses in tomato in the western hemisphere. Plant Dis. 81:1358-1369.
Scott, J. W. 2001. Geminivirus resistance derived from Lycopersicum chilense accessions LA1932, LA1938, and LA2779, in Proceedings of Tomato Breeders Roundtable, Antigua, Guatemala, www.oardc.ohio-state.edu/tomato/TBRT%202001%20Abstracts.pdf.
Scott, J. W. and D. J. Schuster. 1991. Screening of accessions for resistance to the Florida tomato geminivirus. Tomato Genet. Coop. Rep. 41:48-50.
Scott, J. W., M. R. Stevens, J. H. M. Barten, C. R. Thome, J. E. Polston, D. J. Schuster, and C. A. Serra. 1996. Introgression of resistance to whitefly-transmitted geminiviruses from Lycopersicon chilense to tomato, in Bemisia: Taxonomy, Biology, Damage Control, and Management, Gerling, D. and Mayer, R. T., eds., Intercept Ltd, Andover, Hants, UK, 357-367.
Zakay, Y., N. Navot, M. Zeidan, N. Kedar, H. Rabinowitch, H. Czosnek, and D. Zamir. 1991. Screening Lycopersicon accessions for resistance to tomato yellow leaf curl virus - presence of viral DNA and symptom development. Plant Dis. 75:279-281.
Zamir, D., I. Eksteinmichelson, Y. Zakay, N. Navot, M. Zeidan, M. Sarfatti, Y. Eshed, E. Harel, T. Pleban, H. Vanoss, N. Kedar, H. D. Rabinowitch, and H. Czosnek. 1994. Mapping and introgression of a tomato yellow leaf curl virus tolerance gene, Ty-1. Theor. Appl. Genet. 88:141-146.
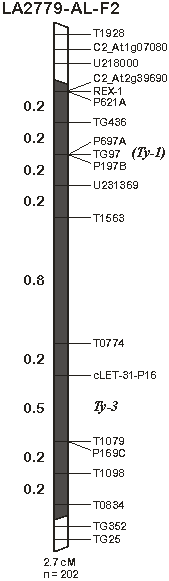
Figure 1. Linkage of the Ty-3 locus to the Ty-1 locus on chromosome 6 of tomato. Linkage maps were constructed from F2 populations of susceptible Solanum lycopersicum × resistant LA2779-derived advanced breeding line. All the markers are PCR-based, including SCAR markers (P621A, P197B, P697A, and P169C) converted from RAPD markers, and CAPS markers taken from either the public domain or designed from the public sequences except T1098 and T0834, which are kindly provided by C. T, Martin and D. P. Maxwell. Shaded regions represented introgression from S. chilense. The markers in non-introgression regions are not drawn to scale.