Evaluation of a co-dominant SCAR marker for detection of the Mi-1 locus for resistance to root-knot nematode in tomato germplasm
Stuart Seah1,4, Valerie M. Williamson1, Brenda E. Garcia2,3, L. Mejía3, Melinda S. Salus2, Christopher T. Martin2, and Douglas P. Maxwell2
1Department of Nematology, University of California, Davis, CA 95616
2Department of Plant Pathology, University of Wisconsin, Madison, WI 53706
3Universidad de San Carlos, Guatemala
4Current address: Commonwealth Scientific and Industrial Research Organisation (CSIRO) Entomology, Private Bag 5, Wembley, WA 6913, Australia
Email: dpmax@plantpath.wisc.edu
The Mi-1 resistance gene was introgressed into cultivated tomato from Solanum peruvianum in the 1940’s (Smith, 1944). This gene confers resistance against many isolates of the root-knot nematode species Meloidogyne incognita, M. javanica, and M. arenaria and is currently the only source of root-knot nematode resistance in modern tomato cultivars. The principle means for developing nematode-resistant tomato cultivars is by traditional breeding aided by marker-assisted selection to detect the Mi-1 gene. Co-dominant CAPS markers such as REX-1 (Williamson et al., 1994) and Cor-Mi (Contact Cornell University Foundation, Ithaca, NY) are widely used to assay for the Mi-1 gene in tomato. Although these markers are generally reliable, El Mehrach et al. (2005) found that both gave false positives for nematode resistance with germplasm derived from Ih902 (F1, F2, Ve), which has begomovirus-resistance genes reportedly introgressed from Solanum habrochaites (listed as 902 in Vidavsky and Czosnek, 1998), but is susceptible to root-knot nematode (V. Williamson, unpublished data). Ih902 is one of the main sources of begomovirus resistance in the tomato breeding program at San Carlos University (Mejía et al., 2005), and it is important to have a breeder-friendly marker that does not give false positive results. Therefore primers were designed that amplify a PCR fragment only if the Mi-1.2 gene, the functional gene for resistance (Milligan et al., 1998), is present. However, these primers do not distinguish homozygous and heterozygous plants (El Mehrach et al., 2005). Here we report evaluation of a co-dominant SCAR marker Mi-23 that is tightly linked to the Mi-1.2 gene.
Material and Methods
Primers: The region on the short arm of chromosome 6 where the Mi-1 locus is located is well characterized genetically and physically (see Seah et al., 2004, 2007). The Mi-1 locus in both resistant and susceptible tomato consists of two clusters with three and four copies of Mi gene homologues, which in resistant tomatoes are separated by approximately 300 kb. Comparison of sequence downstream of Mi-1.2 with a conserved region from susceptible S. lycopersicum led to the development of primers (Mi23F and Mi23R) that flanked an indel within this conserved region (Seah and Williamson, unpublished). The sequence of Mi23F is 5’-TGG AAA AAT GTT GAA TTT CTT TTG-3’, and Mi23R is 5’- GCA TAC TAT ATG GCT TGT TTA CCC-3’.
PCR protocol: DNA was extracted from fresh leaves of plants with PUREGENE® DNA Purification Kit (Gentra Systems, Inc., Minneapolis, MN) and DNA adjusted to approximately 10 ng/µl. PCR was carried out in 25-µl reactions containing 2.5 µl 2.5 mM dNTPs, 5 µl 5X buffer, 2.5 µl 2.5 mM MgCl2, 0.1 µl (0.5 units) GoTaq DNA polymerase (Promega Corp., Madison, WI), 2.5 µl each forward and reverse primer at 10 μM, 2-5 µl of DNA extract, and water. PCR cycles were 94 C for 3 min, the 35 cycles of 94 C for 30 sec, 57 C for 1 min, and 72 C for 1 min. These cycles were followed by 72 C for 10 min, and then the reaction was held at 18 C. PCR reactions were performed in the MJ DNA Engine PT200 Thermocycler™ (MJ Research Inc., Waltham, MA). Amplified fragments were separated by electrophoresis through 2% agarose in 0.5X TBE buffer, then stained with ethidium bromide, and visualized with UV light. For sequencing, ssDNA was digested in PCR reactions with shrimp alkaline phosphatase (Promega Corp.) and exonuclease I (Epicentre, Madison, WI), and the PCR fragments were directly sequenced with Big Dye Sequencing Kit™ and analyzed by the Biotechnology Center, University of Wisconsin-Madison.
Germplasm: The lines M82-1-8 (Ve, F1) and Gh13 were used as the mi/mi genotypes (susceptible); these lines carried the S. lycopersicum sequence for the REX-1 marker (AY596779). Two lines, Motelle and Gh2, which are known to be homozygous for resistance to root-knot nematode (Mi/Mi genotype) have the S. peruvianum sequence for the REX-1 marker (AY729670). The F1 hybrid, Llanero (resistant to begomoviruses, GenTropic Seeds), which is heterozygous for nematode resistance (Mi/mi) (unpublished data), and Marwa (Ve, F2, N and tolerant to Tomato yellow leaf curl virus, Syngenta), which is presumed to be heterozygous for nematode resistance, were used as heterozygous controls. Other commercial F1 hybrids, which were determined to be heterozygous at the REX-1 locus by sequence analysis, were Celebrity (Seminis Seeds), Charanda (Vilmorin), Crista (Harris Moran), Dominique (Hazera Genetics), Tequila (Vilmorin), and Viva Italia (Harris Moran). Rodeo (Heinz) was homozygous at the Mi locus, as determined by the sequence of the REX-1 marker. Titrit (F1, F2, Ve, TMV, FCRR, Royal Sluis) is susceptible to RKN, but is tolerant to Tomato yellow leaf curl virus.
Results and discussion
The susceptible genotypes M82-1-8 and Gh13 (mi/mi) and the resistant genotypes Motelle and Gh2 (Mi/Mi) gave PCR fragments of ca. 430 bp, ca. 380 bp (Fig. 1 or not shown), respectively, as expected for susceptible and resistant tomato. The PCR fragments from M82-1-8 and Gh2 were sequenced and a BLAST search performed at the National Center for Biotechnology Information. The 432-bp fragment (AY596779) from M82-1-8 had 100% nt identity with S. lycopersicum (cv. Heinz 1706, DQ863289) for nt 9,545-9,976, which is located between two resistance-like protein ORFs in cluster 2e. The 377-bp fragment from Gh2 had 100% nt identity with the Mi-1 locus from Motelle (AY729670, S. peruvianum introgression for Mi-1 locus) for nt 25,819-26,195 (U81378), which is located between the Mi-1.2 resistance gene and a pseudo-resistance gene (Mi-1.3) in cluster 1p. Thus, the sequence of the PCR fragments matched the areas of the S. lycopersicum and S. peruvianum genomes used to design the primers. When the two sequences were compared, there were indels of 1 nt and 56 nt, which accounted for the differences in the length of the two sequences. Besides these two indels, there were 13 SNPs between these two sequences.
The heterozygous genotypes Llanero and Marwa gave three fragments, 380, 430 and 500 bp (Fig. 1), respectively. The third, slower moving PCR fragment from the heterozygous plants was hypothesized to be a heteroduplex between the two fragments (380 and 430 bp), which migrated more slowly due to the presence of a 56 nucleotide loop in the heteroduplex molecules. This hypothesis was supported by mixing approximately equal amounts of the 380 and 430-bp fragments and subjecting this mixture to the normal PCR cycle. Three bands of the same size as those from the heterozygous plant resulted from this treatment (Fig. 1, lane 5). When six commercial hybrids (Celebrity, Charanda, Crista, Dominique, Tequila and Viva Italia) with reported resistance to root-knot nematode were tested with the primers Mi23F/Mi23R, all hybrids had the three-banded pattern associated with heterozygous plants for the Mi-1 locus; and Rodeo gave the expected single 380-bp fragment for the homozygous genotype (Mi/Mi). Titrit, which lacked the Mi-1 locus gave the 430-bp fragment corresponding to that associated with the susceptible allele. The Mi23 primers were also tested on 73 breeding lines and hybrids for begomovirus resistance from the tomato breeding project at San Carlos University, Guatemala (Mejía et al., 2005), as well as on 31 other inbreds and hybrids, and unambiguous PCR patterns were obtained.
Previously false positive results for the Mi-1 locus (nematode resistance) were obtained with two co-dominant CAPS markers, REX-1 and Cor-Mi, for the begomovirus-resistant breeding line, Ih902 (El Mehrach et al., 2005). The begomovirus-resistance Ty-1 locus is derived from Solanum chilense LA1969 and was mapped to the short arm of chromosome 6 (Zamir et al., 1994). The REX-1 marker for the TY52 line, which is homozygous for Ty-1/Ty-1 (H. Czosnek, pers. com.), gives a distinct digestion pattern with TaqI restriction enzyme (three fragments, Milo, 2001). Thus, it was of value to test primers Mi23F/Mi23R with tomato lines that gave false positive REX-1 and Cor-Mi restriction digestion patterns, as well as lines known to have the S. chilense introgression for the Ty-1 locus. When Ih902, TY52, and 2 breeding lines (Gc9 and Gc143-2) with the Ty-1 locus were evaluated with the Mi23F/Mi23R primers, only one PCR fragment of 430 bp was obtained. This fragment size was approximately the same as in nematode-susceptible S. lycopersicum, and, thus, the fragment size correlated with the nematode susceptible phenotype of these lines. Sequences of PCR-fragments (EU033925) from the 4 lines were identical, and there were 16 SNPs and a 1-nt indel between this sequence and the sequence from susceptible S. lycopersicum (e.g., M82-1-8). These results indicate that primers Mi23F/R should be adaptable for detecting genotypes with the Ty-1 locus introgressed from S. chilense. This co-dominant SCAR marker has the advantage over previous PCR-based markers in that restriction enzyme digestion of the amplified product is not required, and it does not give false positive fragments with the begomovirus-resistant breeding lines derived from S. habrochaites (Vidavsky and Czosnek, 1998) and S. chilense (Ty-1 locus) (Agrama and Scott, 2006). Additionally, Mi23 may be useful for tomato breeders introgressing other traits located in the resistance gene cluster on the short arm of chromosome 6.
Acknowledgements: This project was funded in part by USAID-CDR (TA-MOU-05-C25-037) and USAID-MERC (GEG—G-00-02-00003-00) grants to D. P. Maxwell, by the College of Agricultural and Life Sciences at University of Wisconsin-Madison and University of California-Davis, and by the US Department of Agriculture’s National Research Initiative Competitive Grants Program (NRICGP; award #00-35300-9410) and the National Science Foundation Award IBN-872 3679 to V. Williamson.
1 2 3 4 5
![]()
![]()
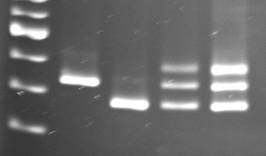
Fig. 1. PCR with primers Mi23F/Mi23R. Lane 1, 100-bp marker (Invitrogen); lane 2, M82-1-8; lane 3, Motelle; lane 4, Llanero (heterozygous), lane 5, equal amounts of the PCR fragments for M82-1-8 and Motelle mixed together and subjected to the standard PCR cycles. Note that three bands are present and that these correspond to the identical sizes of the bands from the heterozygous hybrid Llanero.
References:
Agrama, H.A., and Scott, J.W. 2006. Quantitative trait loci for Tomato yellow leaf curl virus and Tomato mottle virus resistance in tomato. J. Amer. Soc. Hort. Sci. 131:267-272.
El Mehrach, K., Mejía, L., Gharsallah-Couchane, S., Salus, M.S., Martin, C.T., Hatimi, A., Vidavski, F., Williamson, V., and Maxwell, D.P. 2005. PCR-based methods for tagging the Mi-1 locus for resistance to root-knot nematode in begomovirus-resistant tomato germplasm. Acta Hort. 695:263-270.
Mejía, L., Teni, R.E., Vidavski, F., Czosnek, H., Lapidot, M., Nakhla, M.K., and Maxwell, D.P. 2005. Evaluation of tomato germplasm and selection of breeding lines for resistance to begomoviruses in Guatemala. Acta Hort. 695:251-255.
Milligan, S.B., Bodeau, J., Yaghoobi, J., Kaloshian, I., Zabel, P., and Williamson, V.M. 1998. The root knot nematode-resistance gene Mi from tomato is a member of the leucine zipper, nucleotide binding, leucine-rich repeat family of plant genes. Plant Cell 10:1307-1319.
Milo, J. 2001. The PCR-based marker REX-1, linked to the gene Mi, can be used as a marker to TYLCV tolerance. Tomato Breeders Roundtable
www.oardc.ohio-state.edu/tomato/TBRT%202001%20Abstracts.pdf
Seah, S., Yaghoobi, J., Rossi, M., Gleason, C.A., and Williamson, V.M. 2004. The nematode resistance gene, Mi-1, is associated with an inverted chromosomal segment in susceptible compared to resistant tomato. Theor. Appl. Genet. 108:1635-1642.
Seah, S., Telleen, A.C., and Williamson, V.M. 2007. Introgressed and endogenous Mi-1 gene clusters in tomato differ by complex rearrangements in flanking sequences and show sequence exchange and diversifying selection among homologues. Theor. Appl. Genet. 114:1289-1302.
Smith, P.G. 1944. Embryo culture of a tomato species hybrid. Proc. Amer. Soc. Hort. Sci. 44:413-416.
Vidavsky, F., and Czosnek, H. 1998. Tomato breeding lines immune and tolerant to tomato yellow leaf curl virus (TYLCV) issued from Lycopersicon hirsutum. Phytopathology 88:910-914.
Williamson, V.M., Ho, J.Y., Wu, F.F., Miller, N., and Kaloshian, I. 1994. A PCR-based marker tightly linked to the nematode resistance gene, Mi, in tomato. Theor. Appl. Genet. 87:757-763.
Zamir, D., Ekstein-Michelson, I., Zakay, Y., Navot, N., Zeidan, M., Sarfatti, M., Eshed, Y., Harel, E., Pleban, T., van-Oss, H., Kedar, N., Rabinowitch, H.D., and Czosnek, H. 1994. Mapping and introgression of a tomato yellow leaf curl virus tolerance gene, TY-1. Theor. Appl. Genet. 88:141-146.