Occurrence of Anthocyanin in Cultivated Tomato
Peter S. Boches and James R. Myers
Department of Horticulture, Oregon State University, Corvallis, OR 97331 email: myersja@hort.oregonstate.edu, bochesp@hort.oregonstate.edu
Introduction
The cultivated tomato (Solanum lycopersicum) does not normally produce anthocyanins in fruit tissues (Jones et al., 2003). However, in the course of developing a high anthocyanin tomato cultivar at Oregon State University using genes introgressed from wild species, we have noticed several cultivated tomato lines that produce anthocyanin in fruit tissues, especially in green shoulder (u+u+) genotypes. In this article we verify the presence of anthocyanin in these lines using UV-visible spectroscopy.
Materials and methods
Plant materials: These were assembled from various sources and field grown in 2006 and/or 2007 at the Oregon State University Vegetable Research Farm in Corvallis, Oregon (Table 1). Fruit of ‘Purple Smudge’ were grown by Amy Goldman in Rhinebeck, NY and shipped to Oregon for analysis. ‘Purple Smudge’ has enjoyed minor popularity in the heirloom tomato trade, and we only recently learned that it is a Solanum peruvianum (Dun.) introgression line (Young, 1963). A segregating population of anthocyanin fruited seedlings of ‘Dafel’ (an F1 hybrid) were obtained from seed saved from a single plant in a seed lot from Johnny’s Selected Seed Co. that was segregating for plant type and fruit shape. We are unaware of wild species in the pedigrees any of the lines apart from ‘Purple Smudge’.
High anthocyanin breeding lines developed in our program and containing genes introgressed from wild species and non-anthocyanin containing tomatoes were included for comparison as well. The OSU high anthocyanin lines were created by crossing the accessions LA3734A (containing vio, a radiation induced mutant with high anthocyanin stems and veins), LA3736 (containing atv, a gene introgressed from S. cheesmanii LA0434 that causes high anthocyanin in stems and leaves), LA1996 (containing Aft, a gene introgressed from S. chilense that causes anthocyanin accumulation in the fruit), and LA3668 (containing Abg, a gene introgressed from S. lycopersicoides). Although most of our stable high anthocyanin lines (e.g. P20) have LA3668 (Abg) in their pedigree, they probably do not contain Abg, since Abg can not be maintained in a homozygous state due to an inversion on the lower arm of chromosome 10 in S. lycopersicoides relative to S. lycopersicum (Canady et al., 2006).
Anthocyanin Extraction and Measurement
The most highly pigmented fruit from a given line or F2 individual were selected for analysis. For stable breeding lines and named cultivars, each fruit was taken from a separate plant. For F2 individuals, multiple fruit from a single plant were sampled. In 2006, extractions were performed on frozen fruit. In 2007 extractions were performed on fresh fruit. In general, ripe fruit were selected for extraction. However, for ‘Purple Smudge’ and the ‘Dafel’ segregant progeny, extractions were performed on both ripe and green fruit.
Anthocyanins were extracted in acidified methanol using a micro-prep method. Briefly, all pigmented tissue (including the epidermis and some of the pericarp, referred to hereafter as ‘peel’) was removed from single fresh fruit and ground into a fine powder with liquid nitrogen in a mortar and pestle. Fruits were weighed before and after removing the peel to determine the total amount of pigmented tissue. A weighed sub-sample of the homogenized powder was extracted overnight in 300 µl of 1% HCl methanol at 5° C. For the material grown in 2006, a 100-300 mg sub-sample weighed to the nearest 10 mg was used. In order to increase the accuracy of the procedure, in 2007 a 75-150 mg subsample weighed to the nearest 0.1 mg was used. The extraction volume was brought to 500 µl with nanopure water and 500 µl of chloroform was added to the tube. The tubes were centrifuged for 5 minutes at 14,000 rpm and the aqueous phase was removed to a new tube. The presence of anthocyanins was verified by reading absorbance of the samples from 220-750 nm.
Monomeric anthocyanin content was measured using the pH differential method as described by Giusti and Wrolstad (2001). For the pH differential method, the samples were diluted 1:5 with the pH buffer and absorbance was read at an observed λvis-max of 540 nm. The predominant anthocyanin in tomato fruit has previously been reported to be petunidin-3-(p-coumaryl)-rutinoside-5-glucoside (Mes, 2004). For the calculation of the monomeric anthocyanin pigment content, a molecular weight of 934 and a molar absorptivity (ε) of 17000 was used, corresponding to petunidin-3-(p-coumaryl)-rutinoside-5-glucoside in acidified methanol (Price and Wrolstad, 1995). Milligrams anthocyanin per 100 grams of pigmented tissue (peel) on a fresh weight basis was calculated as ((mg/L extract * total volume of extract)/grams peel sampled)*100. Milligrams anthocyanin per 100 grams whole fruit was calculated as (((mg/g peel)*(peel weight in g))/total fruit weight in g)*100.
Results and Discussion
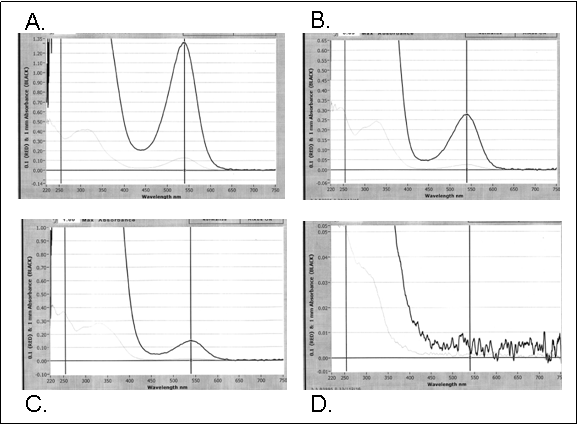
The spectral characteristics of anthocyanin extracts from several tomato cultivars clearly showed that some cultivated tomatoes produce small amounts of anthocyanin in the fruit (Figure 1). In tomato cultivars with very small amounts of visible anthocyanin, such as ‘Yellow Pear’ and ‘Oregon Spring’, an absorbance peak at 540 nm was present but only weakly visible (not shown). In cultivars with modest amounts of anthocyanin such as the ‘Dafel’ segregant progeny (inset C in Figure 1), the peak at 540 nm matched that of the high anthocyanin genotype P20 x hp-2dg (inset A in Figure 1) and clearly differed from the profile of an extract of a tomato with the aa (anthocyanin absent) phenotype (inset D in Figure 1) or that of ‘Legend’ (not shown).
Monomeric anthocyanin contents of the tomato fruit extracts (Table 2) were comparable to those reported by Mes (2004) and Jones et al. (2003) for tomato skin. Mes reported an average value of 13 mg/100 g FW skin for Aft-/atvatv genotypes, and a maximum value of 300 mg/100 g FW skin for Abg-/atvatv genotypes. The value reported by Mes (2004) for Aft-/atvatv skin was lower than that reported by Jones et al. (2003) for Aft skin (~20-60 mg/100 g FW). Mes attributed these differences to the extraction method, which included more of the pericarp tissue in Mes’ extractions. The extraction procedure used here was probably more similar to that of Mes (2004). The value reported here for Aft skin (~30 mg/100 g FW peel) is intermediate between the two. The value reported here for Abg-/atvatv genotypes (84 mg/100g FW peel) is lower than that reported by Mes (2003).
None of the anthocyanin containing cultivars or their segregants had levels of anthocyanin higher than LA1996 (Aft). However, the anthocyanin content of ‘Purple Smudge’ (20 mg/100g FW peel) and the ‘Dafel’ segregant (14.07 mg/100 g FW peel) were close to LA1996 (27.02 mg/100 g FW peel) and the difference was not significant. There was a high coefficient of variation (e.g. ~8-60% for the mean mg/100 g peel in the 2006 data set) in the monomeric anthocyanin content of fruit extracts. This is a common issue in the analysis of plant secondary metabolites (Sumner et al., 2003) and was expected because anthocyanin accumulation in tomato fruit is highly dependent on exposure to light. Sumner et al. (2003) suggest that the average biological variance in plant metabolites for Medicago truncatula is 50%. The lines selected for high anthocyanin accumulation in our program (e.g. P20) typically have a very sparse canopy to allow good light penetration to the fruit. Smaller fruited types (e.g. P4 x ‘Sungold’ small fruited F2) were very high in anthocyanin on a whole fruit basis as a result of their increased surface area:volume ratio, as previously reported by Mes (2003).
The ‘Dafel’ segregant and its progeny are characterized by a very strong green shoulder, sometimes with streaks that descend midway down the fruit. In addition, the green fruit appear to be higher in anthocyanin than ripe fruit, similar to the anthocyanin accumulation pattern observed in some pepper cultivars such as ‘Riot’ or ‘Marbles’. Seed from the ‘Dafel’ segregant was planted in 2007 and the population varied significantly in the amount of green shoulder present on individual plants. Anthocyanin expression was correlated with the degree of green shoulder present, with anthocyanin expression limited to the green shoulder. Anthocyanin expression is limited to the green shoulder in ‘Oregon Spring’ as well. We have also noticed a correlation between green shoulder and anthocyanin expression in material derived from Aft and atv, such as P20. Further work on this topic by our lab may include HPLC analysis of the anthocyanin extracts from tomatoes such as ‘Purple Smudge’, ‘Dafel’ segregants, and ‘Oregon Spring’ and allelism tests between these tomatoes and known anthocyanin fruit genes.
Literature Cited:
Canady M.A., Ji Y., and Chetelat, R.T., 2006. Homeologous recombination in Solanum lycopersicoides introgression lines of cultivated tomato. Genetics 174: 1775-1788.
Giusti M.M. and Wrolstad R.E., 2001. Unit F1.2.1-13. Characterization and measurement of anthocyanins by UV-visible spectroscopy. In: Current Protocols in Food Analytical Chemistry. R.E. Wrolstad (ed). John Wiley & Sons, NY.
Jones C.M., Mes P., Myers J.R., 2003. Characterization and inheritance of the Anthocyanin Fruit (Aft) tomato. Journal of Heredity, 94:449-56.
Mes P., 2004. Breeding tomatoes for improved antioxidant activity. PhD Thesis. Oregon State University.
Price C.L., and Wrolstad R.E., 1995. Anthocyanin pigments of Royal Okanogan Huckleberry juice. Journal of Food Science 60: 369-374.
Sumner L.W., Mendes P., Dixon R.A., 2003. Plant metabolomics: large scale phytochemistry in the functional genomics era. Phytochemistry 62:817-836.
Young, P.A. 1963. Smudged Fruits. Tomato Genetics Cooperative Report 13: 33-34.
Table 1. Plant materials used in this study. Seed Sources are as follows: Johnny’s = Johnny’s Selected Seeds, Winslow, ME; Nichols = Nichols Garden Nursery, Albany, OR; OSU = Oregon State University vegetable breeding program; Amy Goldman = Amy Goldman, pers. comm.; TGRC = Tomato Genetic Resources Center, Davis, CA.
|
Breeding line or cultivar |
Source |
Description |
|
‘Dafel’ segregant and progeny |
Johnny’s |
Anthocyanin containing segregant from ‘Dafel’ and its progeny, strong streaked green shoulder (u+u+), anthocyanin fading somewhat as fruit ripens. |
|
‘Yellow Pear’ |
Nichols |
Uniform ripening (uu) type with occasional anthocyanin in fruit, especially under stress. |
|
‘Legend’ |
OSU |
Anthocyaninless, uniform ripening (uu), OSU developed cultivar. Used as anthocyanin free check in 2006. |
|
‘Oregon Spring’ |
OSU |
Green shoulder (u+u+) OSU cultivar that commonly develops small amounts of anthocyanin in fruit, especially under stress. |
|
‘Purple Smudge’ |
Amy Goldman |
PI 290858, LA2378. S. peruvianum introgression line that develops anthocyanin in crown, a trait that is reportedly without simple Mendelian inheritance. |
|
P13 |
OSU |
Unstable high anthocyanin breeding line developed by crossing Abg x (atv or vio), individuals expressing a strong Abg phenotype were sampled. |
|
P20 |
OSU |
Stable high anthocyanin line developed by crossing [Abg x (atv or vio)] x [atv x Aft], stable F4 individuals were sampled. |
|
P20 x ‘Sweet Baby Girl’ F2 |
OSU |
F2 selection from a P20 x ‘Sweet Baby Girl’ (Seminis) cross. |
|
P20 x LA1194 (‘aa’ F2) |
OSU |
F2 selection from a P20 x LA1194 cross displaying the ‘aa’ (anthocyanin absent) phenotype. |
|
P20 x hp-2dg |
OSU |
F2 individuals from a P20 x (LA3005 x ‘Legend’) cross, selected for hp-2dg and high anthocyanin phenotypes. |
|
P4 x ‘Sunsugar’ |
OSU |
F2 individuals from a (Abg x (atv or vio)) x ‘Sunsugar’ cross. |
|
P4 x ‘Sungold’ |
OSU |
F2 individuals from a (Abg x (atv or vio)) x ‘Sungold’ cross. |
|
P4 x ‘Sungold’ F2 small fruited |
OSU |
F2 individual from a (Abg x (atv or vio)) x ‘Sungold’ cross selected for very small fruit size. |
|
LA1996 (Aft) |
TGRC |
Stock Aft introgression line. |
Table 2. Anthocyanin content of cultivated tomatoes and OSU high anthocyanin breeding lines as measured by the pH differential method. N= number of fruit sampled from separate plants, except for individual F2 plants where multiple fruit were sampled from a single plant; mean mg/100g peel = milligrams anthocyanin per 100 grams of pigmented tissue (peel) on a fresh weight basis; mean mg/100 g whole = milligrams anthocyanin per 100 grams whole fruit on a fresh weight basis; na= not available.
|
Tomato Line or Cultivar |
N |
Mean mg/100 g peel* |
Std dev |
Mean mg/100 g whole* |
Std dev |
|
Field Season 2006 |
|
|
|
|
|
|
P20 |
5 |
111.29a |
8.69 |
3.63a |
0.55 |
|
P13 |
9 |
83.68b |
27.92 |
5.88a |
3.67 |
|
P4 x ‘Sungold’ individual F2 small fruited |
6 |
82.97b |
29.34 |
4.59a |
2.00 |
|
P4 x ‘Sungold’ individual F2 |
6 |
64.79b,c |
8.92 |
5.89a |
1.11 |
|
P4 x ‘Sunsugar’ individual F2 |
6 |
46.67c,d |
23.52 |
4.21a |
2.32 |
|
LA1996 (Aft) |
6 |
27.02d |
11.54 |
0.65b |
0.20 |
|
‘Dafel’ original segregant |
3 |
14.07d,e |
6.09 |
0.24b |
0.08 |
|
‘Oregon Spring’ |
3 |
6.88d,e |
1.75 |
0.13b |
0.01 |
|
‘Yellow Pear’ |
6 |
2.61e |
1.52 |
0.17b |
0.09 |
|
‘Legend’ |
3 |
-0.03e |
0.05 |
0.00b |
0.00 |
|
|
|
|
|
|
|
|
Field Season 2007 |
|
|
|
|
|
|
P20 x hp-2dg, multiple F2 plants |
5 |
84.91a |
34.70 |
6.11a |
3.10 |
|
P20 |
2 |
78.13a |
9.88 |
6.15a |
Na |
|
P20 x ‘Sweet Baby Girl’ individual F2 |
2 |
44.72a |
5.31 |
1.68a |
0.52 |
|
‘Purple Smudge’ breaker fruit |
1 |
20.00a |
Na |
0.15a |
Na |
|
‘Purple Smudge’ green fruit |
2 |
18.12b |
3.14 |
0.30b |
0.06 |
|
‘Dafel’ segregant progeny, green fruit |
1 |
9.47b |
Na |
0.15b |
Na |
|
‘Dafel’ segregant progeny, red fruit |
2 |
1.15b |
0.56 |
0.02b |
0.01 |
|
P20 x LA1194 (‘aa’ F2) |
1 |
0.04b |
Na |
na |
Na |
*Means sharing a superscript not significantly different, Fisher’s LSD, p=0.05.

Figure 1. Spectral characteristics of purified anthocyanins from selected tomato lines. A. P20 X hp-2dg F2 individual, a high anthocyanin (Aft/atv/?Abg), high pigment (hp-2dg) breeding selection, absorbance at 540 nm= 1.303. B.‘Purple Smudge’, an S. peruvianum introgression line, absorbance at 540 nm = 0.278. C. ‘Dafel’ segregant seedling, absorbance at 540 nm = 0.149. D. P20 X LA1194 ‘aa’ F2 segregant, an anthocyaninless tomato, absorbance at 540 nm = 0.003. Note differences in the scale of the absorbance axis for each inset in order to show detail.