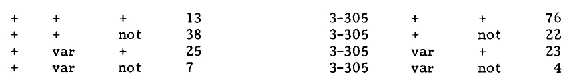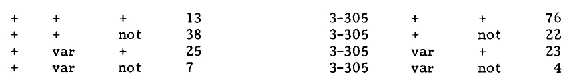Rick, C. M., and P. Bosland Allele tests with
dominants that are homozygous inviable.
Testing for allelism between dominant mutants
that are not viable (i.e., do not reach flowering
stage) poses some difficult problems. Allele
tests are simplest with recessive
mutants because conclusive complementation tests can be made in the F1. With dominant genes that
are homozygous viable, the F2 test is necessary. But with such genes as La, Lpg, Wo, and Xa, no
direct test will work. Heterozygotes between these genes and mimic mutants are presumably
inviable because they have a double dose of the genes in question. Lacking viable heterozygotes
with one dose each of the tester and the new mutation, the essential F2 cannot be produced.
An alternate, indirect approach is to make a linkage test to approximate the locus of the new
mutant. This method yields conclusive results if the new mutant is independent or sufficiently
distant from the tester to yield recombinants; but if no recombinants are obtained, the problem of
tight linkage vs. allelism cannot be resolved.
We recently encountered such a problem with a new Lanceolate-like mutant (3-305) obtained
from Ir. G. J. Hildering of Wageningen, who had induced it in cv. Moneymaker by EMS treatment
and submitted it to the TGC New Mutant Program. This mutant is a near perfect mimic of La in
all observed respects. 1) It is dominant; 2) it is homozygous inviable; 3) the mutant homozygotes
show the same range of phenotypes - "reduced, modified, and narrow" - known in La/La; 4) viability of
the heterozygote is reduced: it appears in subnormal ratios; 5) the heterozygote phenotype is
identical in respect to: precocity, excessive branching, and slender plant parts; although leaves
tend to be unlobed or few-lobed, the extent of lobing exceeds that of +/La. Further, in crosses with
La, no double heterozygotes were obtained, all F2 progenies segregating in the same fashion as for
+/La.
For the linkage test, 3-305 was crossed with our standard linkage tester for chromosome 7 with
the markers var--not. The F2 data are:
The contingency Χ2 between 3-305 and var is 6.25* and that between 3-305 and not is 23.3***.
Clearly, 3-305, like La, is situated somewhere on chromosome 7. The values are not satisfactory
for computing distances; in fact, the great deficiency of 3-305's obstructs the use of any standard
linkage computation. For this kind of situation we prefer to use an empirical curve for the
proportion of the tester gene frequency amongst non-Lanceolate progeny -- for example,
As the crossover distance decreases, the value of this proportion approaches 1.00. When this estimate is
applied to our data, a value of 38 units is estimated for 3-305--var, and 27 units for the 3-305--not
interval. Therefore, to the extent that La lies closer to not than to var, these data suggest that 3-
305 might be allelic with La, but obviously much better tests are needed. We are currently
attempting a testcross for this purpose.
No navigation control above? Click here!